XMapTools documentation for EPMA
Table of contents
EPMA & SEM:
- Data compatibility (EPMA)
- Data conversion
- Generation of mosaics
- Importing data
- Importing calibrated data (EPMA & SEM)
- Classification
- Calibration (EPMA)
Data compatibility for EPMA
CAMECA microprobes
CAMECA Maps
Because of the file format used by CAMECA, XMapTools is reading the information of the following lines in the header:
- Start [map coordinates]
- Step Number [number of columns]
- Line Number [Number of rows]
- Image [to detect the map name].
- If the number of steps and rows doesn't match the size of the table, data won't be imported. The file name is not used to determine the element name when Image is defined.
Therefore the minimum number of header lines is:
Start : X = 15702 Y = -24236 Z = 240
Step Number : 416
Line Number : 440
Y Unit : cts
Image : Ca Ka
[Data table should start here... use tabulation, no coma]XMapTools can import two types of map data file that are generated by CAMECA microprobes.
Type 1: Simple
FileName : Bustamante 18-1-2013.impDat (Current dataset : 4)
Signal(s) Used : Vs1 BSE Z, Na Ka, Ca Ka, Fe Ka, Mg Ka, Al Ka, Si Ka Eds, S Ka Eds, Cl Ka Eds, K Ka Eds, Ti Ka Eds, Mn Ka Eds, Zr La Eds, Ba La Eds
Spectromers Conditions : Sp1 LTAP, Sp2 LPET, Sp3 LLIF, Sp4 LTAP, Sp5 LTAP, Eds 1, Eds 2, Eds 3, Eds 4, Eds 5, Eds 6, Eds 7, Eds 8
Full Spectromers Conditions : Sp1 LTAP(2d= 25.745,K= 0.00218), Sp2 LPET(2d= 8.75,K= 0.000144), Sp3 LLIF(2d= 4.0267,K= 0.000058), Sp4 LTAP(2d= 25.745,K= 0.00218), Sp5 LTAP(2d= 25.745,K= 0.00218), Eds 1, Eds 2, Eds 3, Eds 4, Eds 5, Eds 6, Eds 7, Eds 8
Column Conditions : Cond 1 : 15.1keV 302.5757nA
Date : 19-Jan-2013
User Name : sx
Setup Name : Bustamante.impSet
DataSet Comment :
Comment :
Analysis Date : 1/18/2013 12:42:21 PM
Project Name : Caetano
Sample Name : 2013-01-18
Pha Parameters :
Bias Gain Dtime Blin Wind Mode
Sp1(Na Ka) 1300 2635 3 560 Inte
Sp2(Ca Ka) 1289 1100 3 560 Inte
Sp3(Fe Ka) 1824 550 3 560 Inte
Sp4(Mg Ka) 1293 2964 3 560 Inte
Sp5(Al Ka) 1280 2943 3 560 Inte
EDS: Si Ka
EDS: S Ka
EDS: Cl Ka
EDS: K Ka
EDS: Ti Ka
EDS: Mn Ka
EDS: Zr La
EDS: Ba La
Peak Position : Sp1 46363, Sp2 38388, Sp3 48084, Sp4 38500, Sp5 32460
Start : X = 7386 Y = -17430 Z = 49
Stop :
Dwell Time : 0.03 Sec
Acquisition type : Stage Grid
Step Number : 1001
Line Number : 614
Beam Size : 3 µm
Step Size : 4.
Y Unit :
Image : BSE Z
83.0000 83.0000 88.0000 82.0000 82.0000 81.0000 82.0000 83.0000 83.0000 84.0000 89.0000 92.0000 83.0000 87.0000 83.0000 85.0000 86.0000 86.0000 85.0000 86.0000 88.0000 85.0000 82.0000 87.0000 85.0000 84.0000 87.0000 87.0000 85.0000 85.0000 85.0000 87.0000 86.0000 86.0000 85.0000 86.0000 90.0000 87.0000 87.0000 87.0000 86.0000 96.0000 107.0000 89.0000 88.0000 90.0000 101.0000 113.0000 92.0000 101.0000 81.0000 96.0000 79.0000 81.0000 91.0000 82.0000 128.0000 135.0000 126.0000 120.0000 94.0000 112.0000 84.0000 80.0000 82.0000 83.0000 88.0000 97.0000 82.0000 81.0000 80.0000 81.0000 82.0000 81.0000 131.0000 97.0000 82.0000 83.0000 84.0000 84.0000 80.0000 80.0000 81.0000 87.0000 96.0000 83.0000 84.0000 85.0000 79.0000 82.0000 81.0000 80.0000 81.0000 80.0000 80.0000 79.0000 80.0000 83.0000 82.0000 82.0000 82.0000 82.0000 80.0000 80.0000 86.0000 82.0000 86.0000 85.0000 81.0000 84.0000 83.0000 84.0000 85.0000 83.0000 82.0000 79.0000 81.0000 80.0000 83.0000 84.0000 84.0000 83.0000 84.0000 84.0000 80.0000 80.0000 80.0000 81.0000 85.0000 85.0000 85.0000 83.0000 84.0000 82.0000 83.0000 87.0000 87.0000 84.0000 80.0000 81.0000 85.0000 85.0000 84.0000 83.0000 85.0000 92.0000 81.0000 83.0000 85.0000 85.0000 84.0000 84.0000 84.0000 85.0000 87.0000 87.0000 85.0000 84.0000 84.0000 81.0000 85.0000 80.0000 84.0000 83.0000 84.0000 83.0000 89.0000 91.0000 88.0000 85.0000 89.0000 91.0000 95.0000 91.0000 96.0000 82.0000 84.0000 83.0000 82.0000 86.0000 82.0000 79.0000 86.0000 84.0000 83.0000 83.0000 83.0000 84.0000 83.0000 83.0000 84.0000 84.0000 80.0000 86.0000 86.0000 83.0000 85.0000 83.0000 83.0000 82.0000 91.0000 79.0000 80.0000 76.0000 82.0000 81.0000 81.0000 81.0000 81.0000 81.0000 80.0000 81.0000 83.0000 85.0000 85.0000 86.0000 87.0000 81.0000 83.0000 83.0000 79.0000 80.0000 81.0000 78.0000 84.0000 83.0000 83.0000 76.0000 78.0000 81.0000 80.0000 80.0000 81.0000 80.0000 81.0000 75.0000 81.0000 80.0000 86.0000 115.0000 78.0000 80.0000 83.0000 79.0000 104.0000 102.0000 94.0000 90.0000 84.0000 84.0000 82.0000 84.0000 82.0000 81.0000 81.0000 82.0000 82.0000 83.0000 83.0000 83.0000 83.0000 80.0000 87.0000 98.0000 85.0000 79.0000 82.0000 82.0000 82.0000 83.0000 79.0000 100.0000 115.0000 80.0000 80.0000 80.0000 81.0000 90.0000 75.0000 82.0000 80.0000 79.0000 78.0000 84.0000 81.0000 81.0000 111.0000 107.0000 83.0000 85.0000 83.0000 82.0000 111.0000 84.0000 80.0000 83.0000 70.0000 80.0000 82.0000 83.0000 82.0000 82.0000 81.0000 92.0000 83.0000 86.0000 85.0000 102.0000 101.0000 79.0000 87.0000 84.0000 84.0000 82.0000 86.0000 95.0000 124.0000 84.0000 82.0000 80.0000 85.0000 85.0000 80.0000 81.0000 88.0000 92.0000 88.0000 86.0000 76.0000 83.0000 86.0000 82.0000 82.0000 83.0000 80.0000 80.0000 79.0000 80.0000 80.0000 78.0000 82.0000 88.0000 89.0000 91.0000 88.0000 86.0000 92.0000 97.0000 81.0000 78.0000 79.0000 79.0000 82.0000 82.0000 80.0000 77.0000 80.0000 79.0000 79.0000 80.0000 87.0000 85.0000 85.0000 85.0000 84.0000 83.0000 80.0000 80.0000 79.0000 79.0000 78.0000 84.0000 82.0000 81.0000 80.0000 78.0000 79.0000 80.0000 83.0000 79.0000 81.0000 80.0000 79.0000 77.0000 77.0000 78.0000 85.0000 85.0000 82.0000 85.0000 79.0000 80.0000 80.0000 79.0000 79.0000 79.0000 79.0000 80.0000 85.0000 84.0000 82.0000 80.0000 80.0000 79.0000 79.0000 80.0000 79.0000 80.0000 80.0000 79.0000 80.0000 80.0000 80.0000 82.0000 85.0000 85.0000 85.0000 85.0000 87.0000 80.0000 80.0000 80.0000 84.0000 81.0000 81.0000 94.0000 81.0000 81.0000 81.0000 89.0000 81.0000 80.0000 79.0000 80.0000 85.0000 85.0000 83.0000 84.0000 85.0000 80.0000 80.0000 83.0000 85.0000 88.0000 86.0000 80.0000 80.0000 80.0000 80.0000 80.0000 80.0000 80.0000 81.0000 82.0000 80.0000 83.0000 84.0000 122.0000 99.0000 76.0000 80.0000 82.0000 83.0000 82.0000 86.0000 80.0000 75.0000 91.0000 87.0000 61.0000 74.0000 59.0000 82.0000 94.0000 85.0000 105.0000 87.0000 52.0000 82.0000 88.0000 84.0000 79.0000 80.0000 80.0000 79.0000 80.0000 79.0000 79.0000 81.0000 85.0000 84.0000 84.0000 85.0000 86.0000 85.0000 85.0000 79.0000 80.0000 79.0000 83.0000 80.0000 79.0000 79.0000 79.0000 89.0000 78.0000 80.0000 80.0000 79.0000 79.0000 80.0000 80.0000 81.0000 85.0000 79.0000 78.0000 85.0000 82.0000 78.0000 79.0000 79.0000 78.0000 80.0000 78.0000 78.0000 78.0000 78.0000 79.0000 78.0000 78.0000 78.0000 78.0000 79.0000 79.0000 78.0000 84.0000 85.0000 84.0000 79.0000 79.0000 79.0000 88.0000 83.0000 79.0000 86.0000 98.0000 85.0000 81.0000 80.0000 79.0000 83.0000 79.0000 79.0000 83.0000 85.0000 84.0000 77.0000 77.0000 80.0000 81.0000 79.0000 81.0000 78.0000 78.0000 78.0000 79.0000 78.0000 78.0000 79.0000 79.0000 79.0000 79.0000 86.0000 84.0000 80.0000 79.0000 79.0000 82.0000 80.0000 79.0000 79.0000 78.0000 78.0000 79.0000 79.0000 78.0000 78.0000 79.0000 80.0000 78.0000 85.0000 77.0000 76.0000 78.0000 78.0000 77.0000 76.0000 79.0000 78.0000 79.0000 78.0000 79.0000 84.0000 85.0000 84.0000 85.0000 84.0000 81.0000 85.0000 84.0000 78.0000 78.0000 79.0000 79.0000 78.0000 79.0000 80.0000 80.0000 112.0000 86.0000 88.0000 86.0000 86.0000 84.0000 83.0000 83.0000 84.0000 83.0000 84.0000 76.0000 79.0000 79.0000 78.0000 78.0000 79.0000 79.0000 79.0000 79.0000 79.0000 79.0000 82.0000 89.0000 88.0000 85.0000 85.0000 86.0000 85.0000 84.0000 85.0000 83.0000 85.0000 85.0000 83.0000 89.0000 86.0000 84.0000 86.0000 87.0000 85.0000 86.0000 85.0000 84.0000 87.0000 88.0000 88.0000 85.0000 80.0000 80.0000 80.0000 79.0000 78.0000 79.0000 79.0000 79.0000 80.0000 79.0000 81.0000 86.0000 85.0000 86.0000 86.0000 84.0000 85.0000 84.0000 84.0000 84.0000 91.0000 85.0000 84.0000 83.0000 83.0000 86.0000 88.0000 89.0000 88.0000 86.0000 85.0000 83.0000 84.0000 84.0000 84.0000 84.0000 83.0000 89.0000 79.0000 78.0000 84.0000 81.0000 84.0000 84.0000 83.0000 80.0000 86.0000 83.0000 81.0000 81.0000 80.0000 78.0000 78.0000 81.0000 78.0000 79.0000 123.0000 107.0000 82.0000 83.0000 78.0000 76.0000 78.0000 77.0000 78.0000 78.0000 81.0000 80.0000 75.0000 77.0000 78.0000 78.0000 84.0000 78.0000 79.0000 79.0000 85.0000 85.0000 84.0000 82.0000 84.0000 85.0000 81.0000 85.0000 83.0000 84.0000 84.0000 84.0000 84.0000 84.0000 85.0000 84.0000 85.0000 84.0000 84.0000 84.0000 85.0000 85.0000 90.0000 86.0000 92.0000 86.0000 85.0000 85.0000 85.0000 86.0000 106.0000 84.0000 83.0000 85.0000 85.0000 84.0000 87.0000 108.0000 84.0000 83.0000 84.0000 84.0000 84.0000 88.0000 83.0000 83.0000 83.0000 84.0000 83.0000 85.0000 94.0000 84.0000 85.0000 85.0000 79.0000 78.0000 78.0000 78.0000 78.0000 79.0000 79.0000 79.0000 79.0000 81.0000 86.0000 117.0000 85.0000 84.0000 85.0000 85.0000 83.0000 105.0000 86.0000 86.0000 89.0000 89.0000 88.0000 89.0000 90.0000 84.0000 84.0000 83.0000 77.0000 76.0000 80.0000 82.0000 79.0000 82.0000 88.0000 82.0000 84.0000 83.0000 92.0000 88.0000 78.0000 78.0000 77.0000 78.0000 78.0000 77.0000 80.0000 80.0000 77.0000 77.0000 77.0000 77.0000 77.0000 82.0000 96.0000 83.0000 83.0000 77.0000 77.0000 77.0000 78.0000 77.0000 78.0000 78.0000 78.0000 84.0000 83.0000 84.0000 84.0000 84.0000 84.0000 83.0000 84.0000 84.0000 83.0000 83.0000 84.0000 82.0000 82.0000 83.0000 83.0000 87.0000 82.0000 77.0000 85.0000 76.0000 77.0000 77.0000 78.0000 78.0000 77.0000 77.0000 78.0000 83.0000 84.0000 84.0000 85.0000 82.0000 82.0000 83.0000 82.0000 82.0000 82.0000 83.0000 86.0000 81.0000 82.0000 84.0000 83.0000 83.0000 83.0000 83.0000 77.0000 77.0000 78.0000 77.0000 76.0000 77.0000 79.0000 80.0000 90.0000 83.0000 82.0000 82.0000 82.0000 82.0000 83.0000 85.0000 87.0000 84.0000 82.0000 90.0000 83.0000 89.0000 82.0000 81.0000 83.0000 83.0000 83.0000 83.0000 83.0000 82.0000 80.0000 77.0000 83.0000 82.0000 82.0000 82.0000 83.0000 82.0000 82.0000 82.0000 78.0000 77.0000 77.0000 77.0000 77.0000 77.0000 77.0000 78.0000 78.0000 88.0000 82.0000 82.0000 83.0000 83.0000 84.0000 83.0000 77.0000 77.0000 77.0000 77.0000 77.0000 77.0000 77.0000 82.0000 76.0000 77.0000 79.0000 76.0000 77.0000 78.0000 76.0000 76.0000 77.0000 76.0000 77.0000 77.0000 74.0000 74.0000 77.0000 77.0000 77.0000 77.0000 76.0000 76.0000 77.0000 77.0000 77.0000 76.0000 77.0000 77.0000 76.0000 77.0000 78.0000
82.0000 83.0000 82.0000 80.0000 79.0000 80.0000 84.0000 81.0000 81.0000 82.0000 85.0000 88.0000 86.0000 86.0000 84.0000 83.0000 83.0000 84.0000 83.0000 84.0000 83.0000 85.0000 85.0000 87.0000 86.0000 84.0000 91.0000 91.0000 85.0000 85.0000 85.0000 86.0000 87.0000 89.0000 91.0000 87.0000 94.0000 85.0000 86.0000 90.0000 128.0000 95.0000 110.0000 115.0000 105.0000 105.0000 93.0000 95.0000 102.0000 78.0000 87.0000 85.0000 80.0000 80.0000 88.0000 80.0000 84.0000 128.0000 112.0000 113.0000 102.0000 126.0000 80.0000 80.0000 80.0000 83.0000 85.0000 82.0000 77.0000 79.0000 79.0000 81.0000 81.0000 81.0000 97.0000 90.0000 81.0000 82.0000 83.0000 82.0000 79.0000 82.0000 82.0000 84.0000 87.0000 82.0000 82.0000 84.0000 77.0000 85.0000 80.0000 80.0000 79.0000 80.0000 79.0000 79.0000 82.0000 82.0000 82.0000 81.0000 81.0000 81.0000 81.0000 81.0000 84.0000 82.0000 82.0000 79.0000 79.0000 81.0000 83.0000 83.0000 84.0000 82.0000 78.0000 78.0000 81.0000 80.0000 84.0000 83.0000 83.0000 82.0000 83.0000 81.0000 79.0000 79.0000 79.0000 79.0000 83.0000 83.0000 83.0000 85.0000 85.0000 80.0000 82.0000 84.0000 84.0000 82.0000 81.0000 83.0000 82.0000 83.0000 83.0000 80.0000 83.0000 82.0000 85.0000 83.0000 79.0000 82.0000 83.0000 83.0000 82.0000 85.0000 86.0000 87.0000 84.0000 84.0000 82.0000 82.0000 83.0000 82.0000 81.0000 83.0000 84.0000 84.0000 89.0000 90.0000 81.0000 87.0000 90.0000 89.0000 90.0000 94.0000 89.0000 81.0000 82.0000 82.0000 80.0000 80.0000 84.0000 79.0000 83.0000 82.0000 82.0000 82.0000 82.0000 83.0000 81.0000 95.0000 84.0000 82.0000 83.0000 86.0000 92.0000 84.0000 81.0000 82.0000 82.0000 81.0000 88.0000 78.0000 80.0000 79.0000 81.0000 80.0000 80.0000 80.0000 81.0000 80.0000 80.0000 82.0000 82.0000 79.0000 82.0000 78.0000 83.0000 80.0000 82.0000 83.0000 78.0000 81.0000 80.0000 83.0000 82.0000 82.0000 84.0000 76.0000 78.0000 79.0000 79.0000 80.0000 81.0000 79.0000 80.0000 80.0000 79.0000 80.0000 82.0000 123.0000 88.0000 78.0000 81.0000 75.0000 89.0000 84.0000 90.0000 87.0000 82.0000 160.0000 105.0000 97.0000 82.0000 79.0000 82.0000 81.0000 82.0000 81.0000 82.0000 81.0000 82.0000 75.0000 89.0000 85.0000 83.0000 79.0000 80.0000 81.0000 80.0000 81.0000 82.0000 105.0000 93.0000 79.0000 80.0000 80.0000 80.0000 106.0000 81.0000 77.0000 79.0000 78.0000 83.0000 82.0000 80.0000 82.0000 125.0000 84.0000 83.0000 81.0000 87.0000 81.0000 73.0000 82.0000 82.0000 83.0000 91.0000 81.0000 80.0000 80.0000 81.0000 81.0000 81.0000 87.0000 82.0000 84.0000 82.0000 92.0000 79.0000 78.0000 83.0000 84.0000 82.0000 81.0000 82.0000 87.0000 92.0000 79.0000 79.0000 81.0000 87.0000 79.0000 83.0000 83.0000 86.0000 89.0000 91.0000 79.0000 78.0000 90.0000 78.0000 77.0000 78.0000 83.0000 80.0000 79.0000 79.0000 79.0000 84.0000 79.0000 80.0000 81.0000 83.0000 87.0000 82.0000 79.0000 80.0000 89.0000 81.0000 77.0000 77.0000 80.0000 79.0000 78.0000 80.0000 80.0000 80.0000 78.0000 76.0000 79.0000 82.0000 84.0000 85.0000 84.0000 83.0000 78.0000 78.0000 79.0000 79.0000 79.0000 80.0000 79.0000 78.0000 80.0000 76.0000 77.0000 78.0000 79.0000 79.0000 91.0000 81.0000 79.0000 81.0000 81.0000 82.0000 82.0000 84.0000 84.0000 81.0000 79.0000 82.0000 77.0000 80.0000 80.0000 82.0000 78.0000 78.0000 78.0000 92.0000 81.0000 85.0000 78.0000 84.0000 77.0000 79.0000 79.0000 78.0000 79.0000 79.0000 78.0000 79.0000 86.0000 79.0000 86.0000 84.0000 84.0000 85.0000 85.0000 90.0000 80.0000 79.0000 79.0000 83.0000 80.0000 80.0000 84.0000 79.0000 80.0000 131.0000 91.0000 83.0000 79.0000 80.0000 82.0000 82.0000 85.0000 80.0000 80.0000 80.0000 79.0000 80.0000 81.0000 84.0000 88.0000 86.0000 80.0000 80.0000 80.0000 80.0000 80.0000 79.0000 80.0000 80.0000 81.0000 81.0000 83.0000 86.0000 103.0000 86.0000 82.0000 81.0000 85.0000 88.0000 85.0000 83.0000 86.0000 59.0000 101.0000 91.0000 57.0000 58.0000 66.0000 61.0000 60.0000 102.0000 87.0000 80.0000 56.0000 90.0000 92.0000 85.0000 81.0000 80.0000 80.0000 81.0000 81.0000 79.0000 78.0000 79.0000 86.0000 84.0000 83.0000 83.0000 84.0000 85.0000 84.0000 78.0000 78.0000 78.0000 85.0000 84.0000 79.0000 78.0000 78.0000 79.0000 78.0000 79.0000 79.0000 78.0000 79.0000 78.0000 79.0000 79.0000 84.0000 85.0000 78.0000 84.0000 84.0000 77.0000 78.0000 78.0000 78.0000 76.0000 75.0000 74.0000 76.0000 78.0000 78.0000 77.0000 75.0000 75.0000 81.0000 78.0000 78.0000 77.0000 83.0000 82.0000 82.0000 77.0000 78.0000 77.0000 83.0000 77.0000 78.0000 84.0000 104.0000 84.0000 75.0000 76.0000 78.0000 82.0000 78.0000 79.0000 83.0000 83.0000 84.0000 78.0000 78.0000 78.0000 75.0000 76.0000 76.0000 77.0000 77.0000 77.0000 77.0000 76.0000 77.0000 78.0000 78.0000 80.0000 85.0000 83.0000 84.0000 79.0000 78.0000 78.0000 75.0000 79.0000 78.0000 78.0000 77.0000 77.0000 78.0000 78.0000 78.0000 77.0000 78.0000 77.0000 77.0000 81.0000 77.0000 75.0000 78.0000 78.0000 77.0000 80.0000 79.0000 78.0000 78.0000 77.0000 80.0000 83.0000 84.0000 84.0000 83.0000 105.0000 84.0000 89.0000 83.0000 76.0000 77.0000 77.0000 78.0000 77.0000 79.0000 79.0000 79.0000 146.0000 87.0000 89.0000 84.0000 85.0000 83.0000 83.0000 82.0000 82.0000 83.0000 81.0000 78.0000 76.0000 78.0000 78.0000 77.0000 78.0000 78.0000 78.0000 78.0000 79.0000 79.0000 81.0000 81.0000 82.0000 83.0000 85.0000 85.0000 82.0000 84.0000 84.0000 84.0000 84.0000 85.0000 83.0000 83.0000 80.0000 86.0000 90.0000 88.0000 82.0000 85.0000 85.0000 85.0000 86.0000 85.0000 84.0000 84.0000 82.0000 79.0000 78.0000 78.0000 78.0000 78.0000 78.0000 79.0000 79.0000 85.0000 81.0000 85.0000 85.0000 85.0000 85.0000 83.0000 85.0000 84.0000 83.0000 84.0000 83.0000 84.0000 83.0000 83.0000 84.0000 93.0000 87.0000 85.0000 83.0000 85.0000 83.0000 83.0000 82.0000 87.0000 85.0000 91.0000 85.0000 85.0000 77.0000 78.0000 82.0000 79.0000 81.0000 82.0000 110.0000 77.0000 76.0000 76.0000 77.0000 79.0000 78.0000 78.0000 78.0000 78.0000 78.0000 77.0000 81.0000 102.0000 78.0000 77.0000 77.0000 77.0000 78.0000 77.0000 76.0000 78.0000 77.0000 79.0000 78.0000 77.0000 78.0000 78.0000 76.0000 76.0000 78.0000 78.0000 83.0000 84.0000 79.0000 82.0000 84.0000 84.0000 83.0000 83.0000 84.0000 83.0000 84.0000 84.0000 84.0000 85.0000 83.0000 86.0000 90.0000 83.0000 83.0000 84.0000 85.0000 84.0000 84.0000 83.0000 84.0000 85.0000 85.0000 84.0000 84.0000 95.0000 102.0000 83.0000 83.0000 84.0000 84.0000 82.0000 83.0000 95.0000 83.0000 83.0000 83.0000 89.0000 83.0000 83.0000 82.0000 83.0000 83.0000 82.0000 83.0000 82.0000 85.0000 82.0000 83.0000 83.0000 78.0000 78.0000 77.0000 77.0000 78.0000 78.0000 78.0000 78.0000 78.0000 81.0000 90.0000 89.0000 83.0000 85.0000 85.0000 96.0000 116.0000 101.0000 84.0000 80.0000 90.0000 86.0000 88.0000 87.0000 87.0000 82.0000 82.0000 78.0000 77.0000 88.0000 78.0000 77.0000 76.0000 79.0000 84.0000 82.0000 83.0000 83.0000 81.0000 77.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 88.0000 76.0000 76.0000 77.0000 76.0000 77.0000 76.0000 81.0000 83.0000 85.0000 80.0000 76.0000 77.0000 76.0000 77.0000 76.0000 77.0000 77.0000 78.0000 82.0000 82.0000 83.0000 84.0000 85.0000 83.0000 82.0000 82.0000 82.0000 81.0000 84.0000 83.0000 82.0000 81.0000 82.0000 82.0000 83.0000 82.0000 81.0000 77.0000 78.0000 76.0000 76.0000 77.0000 77.0000 76.0000 77.0000 82.0000 80.0000 82.0000 83.0000 83.0000 82.0000 81.0000 80.0000 82.0000 82.0000 82.0000 81.0000 84.0000 81.0000 82.0000 83.0000 80.0000 80.0000 82.0000 76.0000 76.0000 76.0000 77.0000 77.0000 76.0000 77.0000 78.0000 76.0000 79.0000 82.0000 82.0000 82.0000 82.0000 82.0000 88.0000 85.0000 86.0000 82.0000 82.0000 107.0000 83.0000 85.0000 80.0000 78.0000 83.0000 82.0000 85.0000 81.0000 82.0000 81.0000 78.0000 76.0000 83.0000 82.0000 80.0000 80.0000 81.0000 81.0000 82.0000 81.0000 74.0000 76.0000 76.0000 77.0000 77.0000 76.0000 76.0000 78.0000 77.0000 77.0000 81.0000 82.0000 82.0000 92.0000 83.0000 82.0000 76.0000 79.0000 76.0000 77.0000 77.0000 76.0000 77.0000 81.0000 77.0000 77.0000 80.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 76.0000 77.0000 76.0000 76.0000 77.0000 77.0000Type 2: With column and row numbers
FileName : Celine_091321.impDat (Current dataset : 1)
Signal(s) Used : Ca Ka, Na Ka, K Ka, Al Ka, Mn Ka, Cr Ka, Mg Ka, Ti Ka, Si Ka, Fe Ka
Spectrometers Conditions : Sp1 PET, Sp2 LTAP, Sp3 LPET, Sp4 TAP, Sp5 LLIF, Sp1 PET, Sp2 LTAP, Sp3 LPET, Sp4 TAP, Sp5 LLIF
Full Spectrometers Conditions : Sp1 PET(2d= 8.75,K= 0.000144), Sp2 LTAP(2d= 25.745,K= 0.00218), Sp3 LPET(2d= 8.75,K= 0.000144), Sp4 TAP(2d= 25.745,K= 0.00218), Sp5 LLIF(2d= 4.0267,K= 5.8E-05), Sp1 PET(2d= 8.75,K= 0.000144), Sp2 LTAP(2d= 25.745,K= 0.00218), Sp3 LPET(2d= 8.75,K= 0.000144), Sp4 TAP(2d= 25.745,K= 0.00218), Sp5 LLIF(2d= 4.0267,K= 5.8E-05)
Column Conditions : Cond 1 : 15keV 278.8476nA , Cond 2 : 15keV 280.8923nA
, Cond 1 : Ca Ka, Na Ka, K Ka, Al Ka, Mn Ka
, Cond 2 : Cr Ka, Mg Ka, Ti Ka, Si Ka, Fe Ka
Date : Sep-15-2021
User Name : SX-663\SX-User
Setup Name : C:\SX data\Analysis Setups\Image&Profiles\Celine_091321.impSet
DataSet Comment : BOO17-5N_Map1
Comment :
Analysis Date : Tuesday, September 14, 2021 6:57:42 AM
Project Name : Default Project
Sample Name : Default Sample
Analysis Parameters :
Sp Elements Xtal Position Bias Gain Dtime Blin Wind Mode
Sp1 Ca Ka PET 38388 1304 1031 3 560 Inte
Sp2 Na Ka LTAP 46363 1311 3055 3 560 Inte
Sp3 K Ka LPET 42765 1846 985 3 560 Inte
Sp4 Al Ka TAP 32459 1325 3181 3 560 Inte
Sp5 Mn Ka LLIF 52202 1820 411 3 560 Inte
Sp1 Cr Ka PET 26172 1304 1031 3 560 Inte
Sp2 Mg Ka LTAP 38500 1311 3055 3 560 Inte
Sp3 Ti Ka LPET 31416 1846 985 3 560 Inte
Sp4 Si Ka TAP 27737 1325 3181 3 560 Inte
Sp5 Fe Ka LLIF 48083 1820 411 3 560 Inte
Peak Position : Sp1 38388, Sp2 46363, Sp3 42765, Sp4 32459, Sp5 52202, Sp1 26172, Sp2 38500, Sp3 31416, Sp4 27737, Sp5 48083
Start : X = -1748 Y = -29677 Z = 14
Stop :
Dwell Time : 0.07 Sec
Acquisition type : Stage Grid
Step Number : 504
Line Number : 504
Beam Size : N/A
Step Size : 4.000
Y Unit : cts
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 357 358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392 393 394 395 396 397 398 399 400 401 402 403 404 405 406 407 408 409 410 411 412 413 414 415 416 417 418 419 420 421 422 423 424 425 426 427 428 429 430 431 432 433 434 435 436 437 438 439 440 441 442 443 444 445 446 447 448 449 450 451 452 453 454 455 456 457 458 459 460 461 462 463 464 465 466 467 468 469 470 471 472 473 474 475 476 477 478 479 480 481 482 483 484 485 486 487 488 489 490 491 492 493 494 495 496 497 498 499 500 501 502 503 504
1 1987 1954 2079 1974 2083 2103 2073 2128 2028 1850 2121 1916 1938 2013 2066 2037 2109 2075 2093 2032 2053 2087 2118 2130 2082 2047 1986 2056 1994 2006 1920 1949 1970 1981 1922 2279 1986 1984 1992 1983 1960 1958 1996 1910 1972 1939 1934 1962 1984 1948 1933 1929 1850 1921 1948 1941 1942 1978 1997 2014 1973 1939 1985 1871 1595 1935 1964 1864 1905 1894 1896 1836 1894 1908 1920 1804 1876 1871 1812 1833 1928 1830 1844 1874 1982 1853 1934 1857 1920 1837 1856 1882 1964 2920 2025 1817 1908 1967 1925 1901 1947 1895 1912 1886 1967 1898 1945 1939 1935 1975 1962 2007 1963 1930 1941 1914 1893 1994 1986 1930 1954 2039 1950 2004 2005 1976 1943 1906 2046 2070 2105 2129 2971 2420 2304 2475 2051 2036 2184 2213 2047 2049 2064 2043 2788 2104 1979 1972 2056 2022 2153 2087 2180 2124 2094 1993 1992 2097 1957 2016 2014 3758 2197 2042 2032 2024 2018 2226 2570 3155 2870 2174 2000 1983 1942 2018 1974 1944 2043 1962 2047 1954 1961 2035 2083 2085 2003 2020 2069 2153 1988 1957 2014 2087 2054 1963 2052 2043 1999 1947 2007 2109 2035 2016 2100 2000 2013 1980 2012 2051 2072 2083 1978 2015 2078 2232 3343 3864 3847 2105 3254 3823 3869 3952 4521 3751 4286 4042 3589 3693 3570 3397 3597 3553 3877 3837 3747 3997 3824 2952 2986 3676 3975 4050 4008 3699 3649 3843 3940 4099 3524 3987 4367 3461 2725 2960 2424 2145 2095 2132 1941 2070 2081 2488 2146 2270 2069 2080 2078 2062 2666 2099 2120 2152 2076 2052 2041 2033 2054 2067 2222 2012 2114 2088 2031 2142 2107 2265 2502 2852 3239 3536 3372 3914 2630 3431 3709 2810 3996 3249 3964 2999 3846 3832 3698 3362 2881 2706 2166 1969 1923 2051 2080 2138 2002 2054 2021 1992 2047 1967 2025 2007 1934 2068 1980 1990 1914 2039 1982 1995 2016 2098 2067 2048 1999 2044 3706 3266 1668 1885 1802 2099 1990 2050 1939 2059 2032 2039 2112 2146 2052 2088 2062 2041 2006 2086 2084 2090 2087 2046 1996 2078 2103 2030 2069 2018 2019 1994 1957 2042 1978 2160 2823 3063 2137 2088 2327 1857 2111 2065 2149 2063 2049 1962 1987 2056 2027 2041 1925 1945 2027 2001 1985 2006 2000 2033 1998 1951 1929 1985 1983 1939 1983 1967 1990 2063 1946 2017 1998 1990 2039 2048 2012 1996 2065 2041 2001 2053 2022 2013 1974 2074 2572 2105 2021 2040 2064 2071 2022 2020 2057 2032 2056 2095 1877 2073 1991 1990 2428 1931 2029 2031 2016 2006 2046 2024 1984 1941 2071 2061 2049 2042 1974 2010 1989 1970 2000 1984 2010 2001 1932 1921 1949 1958 2033 2998 1998 2039 2012 2023 1988 2020 2522 2108 1998 2059 2051 2008 2054 2096 2009 1914 1802 1440 1605 1981 2049 1930 2037 1900 1945 1919 1976 1969 1966 1857 1934 1934 1913 1958 1862 1945 1895 1943
2 2332 2198 2226 2202 2107 2090 2033 2026 1757 2032 2119 1966 1903 2029 2090 2051 2144 1991 1874 2096 2019 2018 2093 2067 2154 1942 1989 2095 1990 2047 1987 2005 1986 1939 2445 2190 2034 2024 1979 1945 1995 1999 1934 2001 1962 1955 1965 1918 1902 1908 1922 1963 1921 1935 1928 1966 1953 1925 1986 1952 1993 1934 2000 1935 1301 1842 1905 1886 1910 1884 1861 1874 1887 1869 1871 1802 1881 1864 1846 1880 1891 1838 1866 1829 1805 1804 1908 1856 1972 1823 1903 1921 2755 1959 1915 1884 1873 1890 1936 1924 1959 2005 1863 1956 1926 1949 1909 2007 1982 1855 1999 1905 1855 1917 1921 1975 1917 2006 1979 1977 2020 2010 1995 1968 1944 1922 1949 1987 2004 2045 2096 2131 2339 2289 2020 2457 2116 1978 2056 2164 2286 2044 2068 2084 2587 2245 1952 1826 1988 2122 2205 1928 2096 2107 2095 2072 2077 1989 2030 1904 1963 3104 1961 2092 2102 2375 3134 3825 4043 3994 3763 3068 2383 1956 1995 2063 1986 1965 1923 2017 2094 2013 2049 2000 2006 2047 2078 2063 1979 2092 2001 1960 2014 2033 2011 2005 2002 1992 2011 1993 2067 2034 1982 2026 2088 2035 2025 2038 2055 2065 2005 2093 2043 1990 2157 2078 2801 3693 3767 3727 2846 3401 4214 4116 3366 3732 3929 4034 3755 3730 3353 3589 3412 3894 4008 4006 3844 4042 3954 3862 3095 4028 4009 3864 4144 3949 3285 3684 2972 3954 4209 3549 3351 3483 3512 2830 3129 2177 2469 2111 2167 2063 2209 2344 2205 2145 2187 2107 1978 1984 2621 2051 2018 1989 2032 2002 2061 2105 2081 2265 2051 2040 2101 2085 2115 2125 2868 3459 3643 3598 3684 3661 3244 3541 3935 3975 4090 4612 3863 3959 3461 3508 3268 3851 3696 3301 2613 2099 1899 1983 2001 2060 1986 2044 2080 1987 2019 2083 2025 1916 1930 2083 1962 2085 2002 2009 2010 2044 2010 1980 2011 2021 2049 2040 2040 2096 2394 4142 3000 1991 2024 1767 2041 2045 1954 2069 2093 1933 2081 2131 2158 2050 2027 1994 2154 2091 2202 2044 2086 2057 2024 2064 1950 2045 2007 2068 1990 1992 1937 1974 1964 2363 3343 2345 2163 2123 2050 1155 2049 2024 2059 2012 1987 2073 2126 2034 1927 2006 1949 1967 1983 1987 1990 1983 2123 1999 1909 1939 1982 1976 1991 1950 1969 2074 1954 2042 1938 2043 2038 2022 1970 2005 1995 2101 1987 2027 2063 2035 1972 1972 1955 1939 2440 2033 2080 2033 2086 2038 2053 2037 2047 2043 2110 2024 1955 2027 1997 1854 2383 2014 2019 2018 2114 2001 2047 2003 2030 2040 2086 2043 2020 1996 1996 2041 1976 1904 1922 1964 2012 1971 2012 1930 1940 2013 3141 2025 1982 2039 1976 1957 1920 2018 2133 2033 2096 1982 2105 1929 1965 2043 1920 1806 2002 1985 1959 2016 1909 1995 1971 1986 1964 1914 1917 1942 1956 1921 1917 1927 1920 2006 1977 1948 1875 1857Spot analyses
XMapTools can import two types of spot data file that are generated by CAMECA microprobes.
Type 1: Oxide only
DataSet/Point Na2O MgO SiO2 Al2O3 K2O FeO MnO BaO Cr2O3 Cl CaO TiO2 Total X Y Z Comment Mean Z Date
1 / 1 . 0.000 0.047 0.000 0.000 0.012 0.049 0.102 0.000 0.000 0.019 54.334 0.000 54.562 7160 -17670 49 9.049 21.01.13 12:56
2 / 2 . 0.043 0.020 29.964 1.474 0.019 0.716 0.118 0.435 0.014 0.013 27.921 37.349 98.086 6456 -17357 50 14.564 21.01.13 12:59In this example data from Na2O to TiO2 are imported.
Each line represents a single analysis. The first entry DataSet/Point is defined using space (noted ” ” below) whereas tabulations (“\t”) are used to separate other entries. Only this format can be properly read by XMapTools. The correct format for each data row is:
Number” “/” “Number” “.”\t”Number”\t”Number”\t”Number …Example:
DataSet/Point Na2O MgO SiO2 Al2O3 K2O FeO MnO BaO Cr2O3 Cl CaO TiO2 Total X Y Z Comment Mean Z Date
1 / 1 . 0.000 0.047 0.000 0.000 0.012 0.049 0.102 0.000 0.000 0.019 54.334 0.000 54.562 7160 -17670 49 9.049 21.01.13 12:56
2 / 2 . 0.043 0.020 29.964 1.474 0.019 0.716 0.118 0.435 0.014 0.013 27.921 37.349 98.086 6456 -17357 50 14.564 21.01.13 12:59Alternatively, the following input also works:
DataSet/Point Na2O MgO SiO2 Al2O3 K2O FeO MnO BaO Cr2O3 Cl CaO TiO2 Total X Y Z Comment Mean Z Date
1 0.000 0.047 0.000 0.000 0.012 0.049 0.102 0.000 0.000 0.019 54.334 0.000 54.562 7160 -17670 49 9.049 21.01.13 12:56
2 0.043 0.020 29.964 1.474 0.019 0.716 0.118 0.435 0.014 0.013 27.921 37.349 98.086 6456 -17357 50 14.564 21.01.13 12:59Type 2: Full output
An example is provided below. In this example, data ranging from Na₂O to FeO is imported.
FileName : Celine_test_091321.qtiDat
Signal(s) Used : Na Ka, Mg Ka, Al Ka, Si Ka, K Ka, Ca Ka, Ti Ka, Cr Ka, Mn Ka, Fe Ka
Spectrometers Conditions : Sp2 LTAP, Sp2 LTAP, Sp4 TAP, Sp4 TAP, Sp3 LPET, Sp1 PET, Sp3 LPET, Sp1 PET, Sp5 LLIF, Sp5 LLIF
Full Spectrometers Conditions : Sp2 LTAP(2d= 25.745,K= 0.00218), Sp2 LTAP(2d= 25.745,K= 0.00218), Sp4 TAP(2d= 25.745,K= 0.00218), Sp4 TAP(2d= 25.745,K= 0.00218), Sp3 LPET(2d= 8.75,K= 0.000144), Sp1 PET(2d= 8.75,K= 0.000144), Sp3 LPET(2d= 8.75,K= 0.000144), Sp1 PET(2d= 8.75,K= 0.000144), Sp5 LLIF(2d= 4.0267,K= 5.8E-05), Sp5 LLIF(2d= 4.0267,K= 5.8E-05)
Column Conditions : Cond 1 : 15keV 20nA
Date : Sep-15-2021
User Name : SX-663\SX-User
Setup Name : C:\SX data\Analysis Setups\Quanti\Celine_091321.qtiSet
DataSet Comment : BOO17-3S_map2_cpx1
Comment :
Analysis Date : Monday, September 13, 2021 4:45:45 PM
Project Name : Default Project
Sample Name : Default Sample
Analysis Parameters :
Sp Elements Xtal Position Bg1 Bg2 Slope Bias Gain Dtime Blin Wind Mode
Sp2 Na Ka LTAP 46369 -700 800 1311 3117 3 560 Inte
Sp2 Mg Ka LTAP 38513 -1150 1150 1311 3055 3 560 Inte
Sp4 Al Ka TAP 32464 800 1.2 1324 3227 3 560 Inte
Sp4 Si Ka TAP 27737 750 1.1 1325 3181 3 560 Inte
Sp3 K Ka LPET 42762 -600 600 1845 981 3 560 Inte
Sp1 Ca Ka PET 38391 700 1.1 1307 1026 3 560 Inte
Sp3 Ti Ka LPET 31406 600 1.05 1846 985 3 560 Inte
Sp1 Cr Ka PET 26186 -500 500 1304 1031 3 560 Inte
Sp5 Mn Ka LLIF 52200 650 1.05 1820 412 3 560 Inte
Sp5 Fe Ka LLIF 48084 800 1.2 1820 411 3 560 Inte
Peak Position : Sp2 46369 (-700, 800), Sp2 38513 (-1150, 1150), Sp4 32464 (800, Slope = 1.2), Sp4 27737 (750, Slope = 1.1), Sp3 42762 (-600, 600), Sp1 38391 (700, Slope = 1.1), Sp3 31406 (600, Slope = 1.05), Sp1 26186 (-500, 500), Sp5 52200 (650, Slope = 1.05), Sp5 48084 (800, Slope = 1.2)
Current Sample Position : X = -10413 Y = 32015 Z = 60 BeamX = 0.00 BeamX = 0.00
Standard Name :
Na ,Al On albite
Mg ,Si ,Ca On Wakefield diopside
K On kspar
Ti On TiO2
Cr On MgCr2O4
Mn On Rhodon 41522 AMNH
Fe On RKFAYb7
Standard composition :
albite = Na : 8.77%, Al : 10.29%, Si : 32.13%, O : 48.81%
Wakefield diopside = Si : 25.94%, O : 44.43%, Na : 0.02%, Mg : 11.22%, Al : 0.02%, Ca : 18.53%, Ti : 0.02%, Mn : 0.02%, Fe : 0.09%
kspar = O : 45.94%, Na : 0.85%, Al : 9.83%, Si : 30.10%, K : 12.39%, Ba : 0.70%
TiO2 = Ti : 59.95%, O : 40.05%
MgCr2O4 = Mg : 12.64%, Cr : 54.08%, O : 33.28%
Rhodon 41522 AMNH = O : 37.73%, Mg : 2.36%, Si : 21.98%, Ca : 1.06%, Mn : 35.01%, Fe : 1.80%
RKFAYb7 = Si : 13.84%, O : 31.37%, Mg : 0.06%, Al : 0.05%, Ca : 0.02%, Mn : 1.55%, Fe : 52.62%, Zn : 0.38%
Calibration file name (Element intensity cps/nA) :
Na ,Al : Other\albite_15kV_NaKa-Sp2-LTAP_AlKa-Sp4-TAP_010.calDat (Na : 127.8 cps/nA, Al : 189.2 cps/nA)
Mg ,Si ,Ca : Other\Wakefield diopside_15kV_MgKa-Sp2-LTAP_SiKa-Sp4-TAP_CaKa-Sp1-PET_019.calDat (Mg : 315.4 cps/nA, Si : 541.5 cps/nA, Ca : 101.3 cps/nA)
K : Other\kspar_15kV_KKa-Sp3-LPET_017.calDat (K : 208.0 cps/nA)
Ti : Other\TiO2_15kV_TiKa-Sp3-LPET_029.calDat (Ti : 1589.2 cps/nA)
Cr : Other\MgCr2O4_15kV_CrKa-Sp1-PET_013.calDat (Cr : 247.4 cps/nA)
Mn : Other\Rhodon 41522 AMNH_15kV_MnKa-Sp5-LLIF_020.calDat (Mn : 192.7 cps/nA)
Fe : Other\RKFAYb7_15kV_FeKa-Sp5-LLIF_057.calDat (Fe : 317.7 cps/nA)
Beam Size : N/A
Weight% Atomic% Oxide
DataSet/Point Na Mg Al Si K Ca Ti Cr Mn Fe O Total Na Mg Al Si K Ca Ti Cr Mn Fe O Total Na2O MgO Al2O3 SiO2 K2O CaO TiO2 Cr2O3 MnO FeO Total X Y Z Beam X Beam Y Comment Distance (?) Mean Z Point# Date
1 / 1 . 0.743369 7.000587 0.992360 23.675010 0.000010 15.496050 0.028793 0.050500 0.047470 7.429382 41.095420 96.558950 0.753605 6.712945 0.857190 19.646370 0.000006 9.010886 0.014010 0.022636 0.020138 3.100467 59.861750 100.000000 1.002046 11.609060 1.875058 50.649700 0.000012 21.682070 0.048028 0.073809 0.061296 9.557870 96.558950 -10413.0 32015.0 60.0 BOO17-3S_map2_cpx1 0.00 12.714160 1 Monday, September 13, 2021 4:45:45 PM
1 / 2 . 0.683164 7.306072 1.054231 24.247130 0.005349 15.870280 0.049491 0.006556 0.033462 7.281908 42.080150 98.617790 0.676747 6.845810 0.889827 19.661410 0.003116 9.017649 0.023530 0.002872 0.013871 2.969490 59.895680 100.000000 0.920890 12.115650 1.991962 51.873660 0.006443 22.205690 0.082554 0.009582 0.043208 9.368145 98.617790 -10408.3 32015.0 60.0 BOO17-3S_map2_cpx1 4.68 12.939130 2 Monday, September 13, 2021 4:48:12 PMData conversion for EPMA
The EPMA converter can be used to convert raw data for supported instruments to XMapTools format.
The following data formats are currently supported:
- JEOL (WIN) JEOL microprobes running on WINDOWS.
- JEOL (SUN) JEOL microprobes running on SUN-OS.
- CAMECA for recent CAMECA microprobes (see format description).
EPMA converter module
The converter can be accessed via Project and Imports, using the Open XMapTools' EPMA Converter button.
Main steps:
- Select the data format (e.g. JEOL or CAMECA; check compatible data formats: CAMECA).
- Select the destination folder, usually an empty folder where data with the XMapTools format will be stored.
- Select maps which are stored in a given folder; text or csv map files generated by the microprobe software.
- Validate map selection.
- Add standards data exported by the microprobe software as text or csv files. This step can be repeated until all standards data have been imported.
- Generate Standards.txt containing the map coordinates and the spot analysis data to be used as internal standards for map calibration.
For more detailed information on the EPMA Converter, refer to the embedded documentation available within the program.
Step 1: Select data format
Use the Select the format dropdown menu to select data format. After selecting the data format two text boxes are displayed indicating the types of files required for the selected format. The first on the left is for the map and the second one for the spot analyses.
- JEOL (WINDOWS) requires two files for each map: 'data00X.cnd' and 'data00X.csv', and a single file for each set of spot analyses: 'summary.csv'.
- JEOL (SUN) requires two files for each map: 'XX.cnd' and 'XX_map.txt', and two files for each set of spot analyses: 'summary.txt' and 'stage.txt'.
- CAMECA requires one file for each map: 'XXXX.txt', and a single file for each set of spot analyses: 'YYY.csv'.
Step 2: Set the destination folder
After selecting the data format, press the Set a destination folder button and select an empty folder to save the data in the XMapTools format. This folder must be empty and existing data will be deleted!
Note that this folder can then be transferred to a working data folder for separate storage from the raw data.
Step 3: Import maps
After selecting the destination folder, press the Import Maps button to select the folder containing the map files. XMapTools will convert all the maps available in this folder.
After selecting the maps, press the Validate map selection button to continue.
Step 4: Import spot analyses (internal standards)
After selecting all files for the spot analyses, press the button generate Standards.txt. This will end the procedure and close the Converter. Note that when you return in XMapTools, the working directory has been adjusted and you can start importing your maps.
Importing data using the import module
To import map data in XMapTools, select the 'Project and Import' tab and press the Import Maps ![]() button located in 'Import Maps and Images'. This will open the import module and prompt you to select files.
button located in 'Import Maps and Images'. This will open the import module and prompt you to select files.
Select the set of map files you want to import. Choose compatible files from the 'Pick Map File(s)' pop-up window. Note that multiple files can be selected at once. Any selected file that cannot be imported due to an incompatible format, for example, will be skipped during import.
Format: Map files must have the *.txt, *.asc, *.dat or *.csv extension, no header and a name compatible with the XMapTools default element names for identification. The default lists of compatible element and oxide names are given in the source of the help file below.
Selected map files are listed in the main table. More maps can be added by pressing the map selection button (see above)
File: Contains the file names
- Map Name: Contains the name of the corresponding element in the database
- Type: Element or oxide
- Data: Intensity or wt%
- Special: EDS or WDS(?); in the case of WDS(?) a DTC is automatically selected
- DTC: Dead time correction
- OC: Orientation correction (legacy function from XMapTools 3, do not use; not tested)
- Destination: Destination in XMapTools, can be: Intensity, Quanti, Merged, Other; drop down menu, can be edited.
- Action: Keep or eliminate; drop down menu, editable.
- Settings, such as setting the dwell time (as in XMapTools 3) can be changed in the Corrections section.
Press the Import data button to import the selected maps into XMapTools after the corrections have been applied.
For more detailed information on the EPMA Converter, refer to the embedded documentation available within the program.
Importing calibrated data from EPMA and SEM
This section describes how to work in XMapTools with data that have been calibrated by an other program.
The calibrated maps should be translated first into a compatible format (e.g. txt or csv file). Use the element or oxide abbreviation as file name (e.g. Si.txt, SiO2.txt, Ce.txt, etc.).
Step 1: Adding calibrated maps
Quantitative maps can be imported in XMapTools via the Import Tool. Quantitative maps can be expressed in µg/g or wt% of elements (e.g. Si.txt, Al.txt, etc.) or oxides (e.g. SiO2.txt, Al2O3.txt, etc.). These maps are imported in the category 'Merged' data. The data format is not specified during the import.
- For maps expressed in µg/g or wt% of oxides, the destination is automatically set to 'Merged'. No action is required, simply press the button Import Data.
- Maps expressed in µg/g or wt% of elements, the destination must be changed manually to 'Merged' (see figure below). This operation needs to be repeated for each map. The click on the button Import Data.
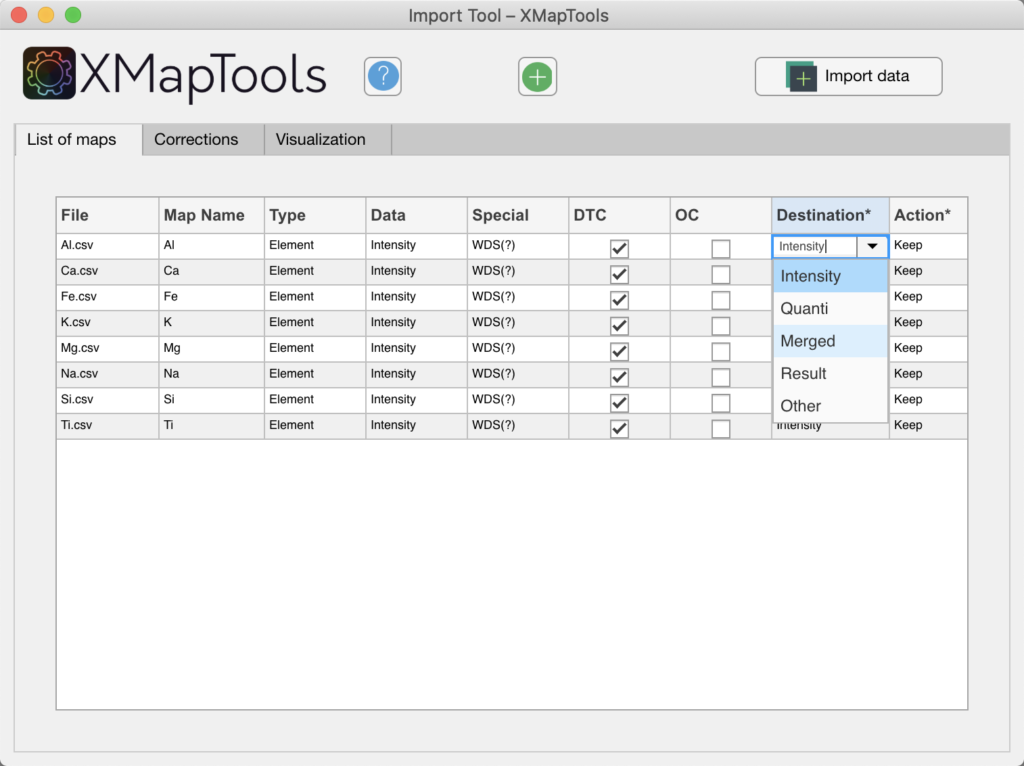
Figure: If maps are expressed in mass of elements, the destination should be changed from 'Intensity' to 'Merged'.
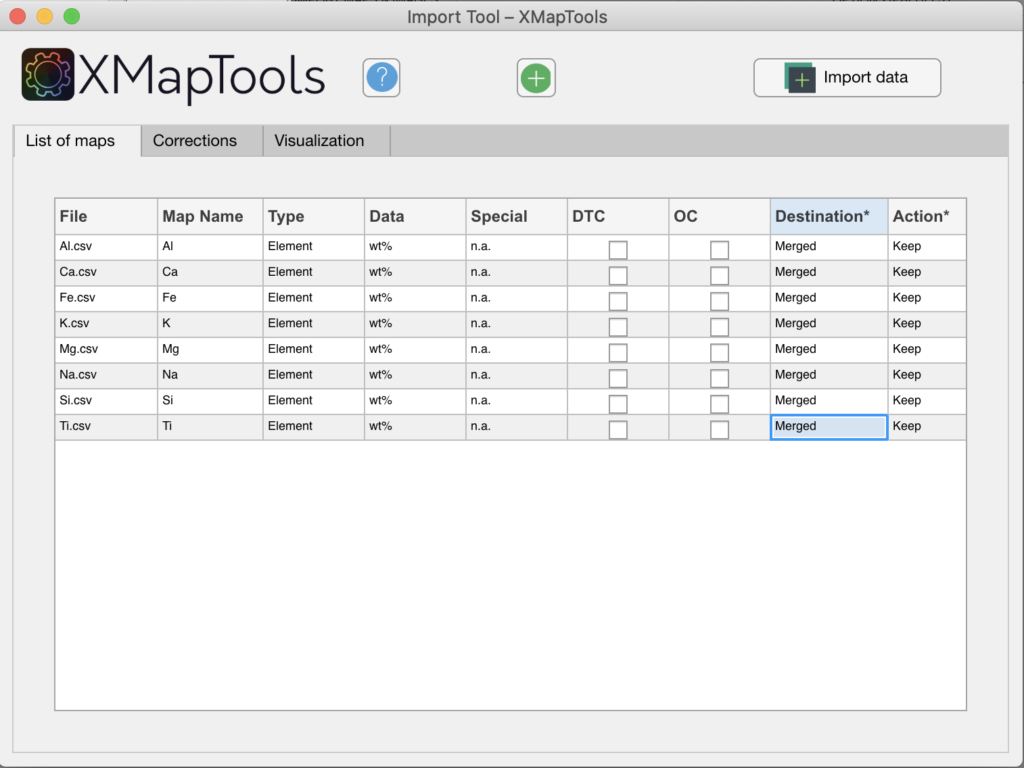
Figure: In this example the destination for each map has been set to 'Merged'.
Step 2: Data conversion (optional)
The compositional maps should ideally be expressed in oxide wt%. if the imported maps are in µg/g or wt% of elements, a conversion step can be required to calculate structural formulas.
In the primary menu, select the dataset Imported_Maps and then right-click on its name. Select the option Convert to open the Converter module.
Select the conversion method in the Converter tool and press the Apply button.
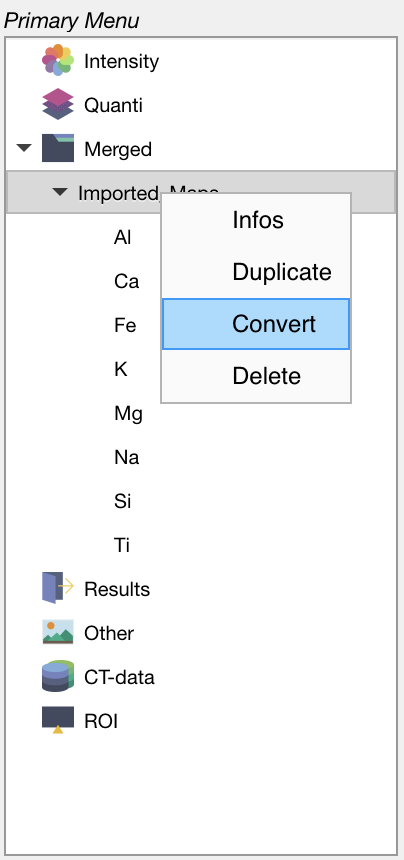
Figure: Select a dataset and then right-click to open the Converter from the primary menu.
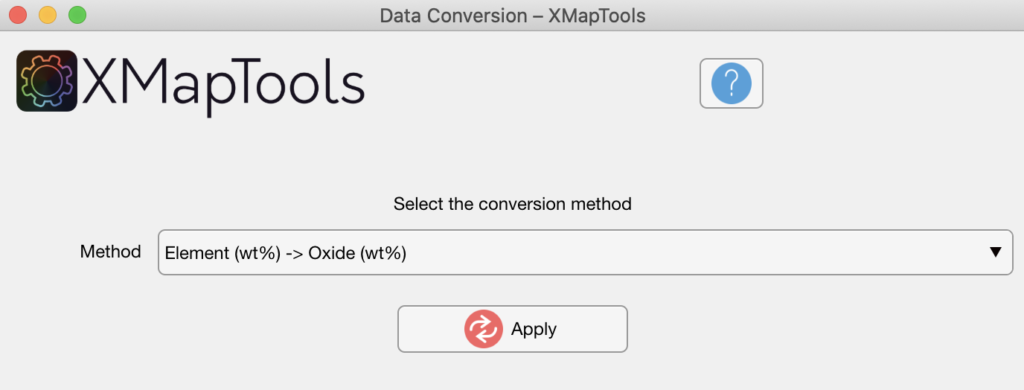
Figure: Data conversion tool. In this example data are converted from element wt% to oxide wt%.
Step 3: Classification of quantitative data
Step 3.1: Create a training set
Display a map from the imported dataset using the Primary Menu and open the Classify tab. Select Training Set (Classification) in the Secondary Menu and press the Add ![]() button in Classify and select the phases. Press again the Add
button in Classify and select the phases. Press again the Add ![]() button to create a new mask definition in the training set. This operation can be repeated until the correct number of phases is reached. Each mask definition can be deleted by right-clicking on the name and selecting Delete.
button to create a new mask definition in the training set. This operation can be repeated until the correct number of phases is reached. Each mask definition can be deleted by right-clicking on the name and selecting Delete.
You can rename each mask definition by double-clicking on his name in the Secondary Menu. Press Add ![]() in Classify when a mask definition is selected to add a region-of-interest (ROI).
in Classify when a mask definition is selected to add a region-of-interest (ROI).
Step 3.2: Add maps for classification
Select the dataset in the category Merged of the Primary Menu and press the Add Maps for Classification ![]() button to add all the maps of the dataset in the list that will be used by the classification function.
button to add all the maps of the dataset in the list that will be used by the classification function.
Step 3.3: Classification
Select a dataset in the Merged category of the Primary Menu and a Training Set in the Secondary Menu. Pick an algorithm in the tab Classify and press the Classify ![]() button. Note that the Classify button is only available when an appropriate dataset and training set are selected in the primary and secondary menus.
button. Note that the Classify button is only available when an appropriate dataset and training set are selected in the primary and secondary menus.
Step 4: Splitting a merged dataset using a maskfile
The imported dataset available in the Merged data category has to be divided into maps for each mask that will be stored under the Quanti data category.
Select a mask file in the Secondary Menu. Select the dataset in Merged and then right-click on its name. Select the Split (using maskfile) option.
The results are stored under the Quanti category as individual dataset, one for each mask. These data sets can be used to calculate maps of structural formulas or other calculations.
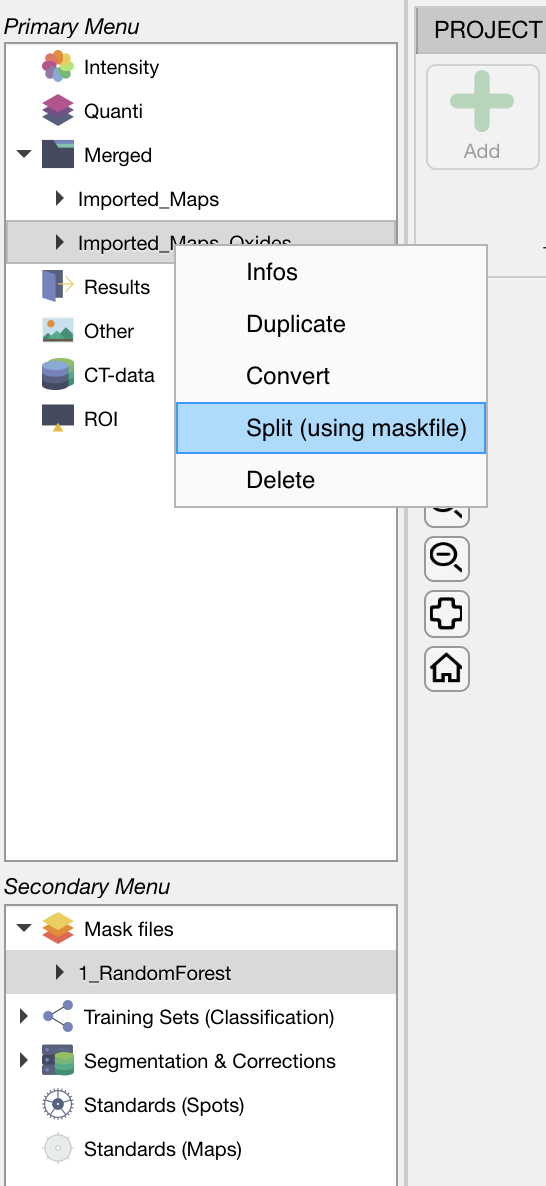
Figure: Select a mask file, then click on the dataset of interest (here Imported_Maps_Oxides) and then right-click on the name for accessing the menu.
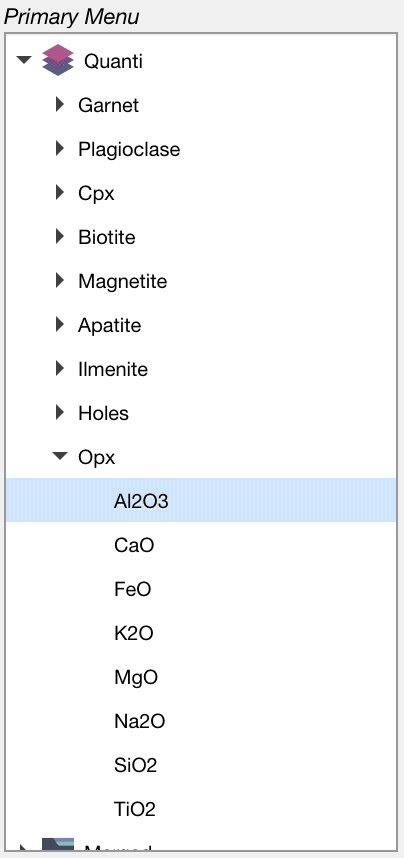
Figure: Results are stored in the category Quanti
Classification
Compositional map classification is the process of categorising and labelling groups of pixels within a dataset based on their composition. It generates a mask image showing the distribution of each mask/class (i.e. features can be mineral/epoxy/glass, etc.).
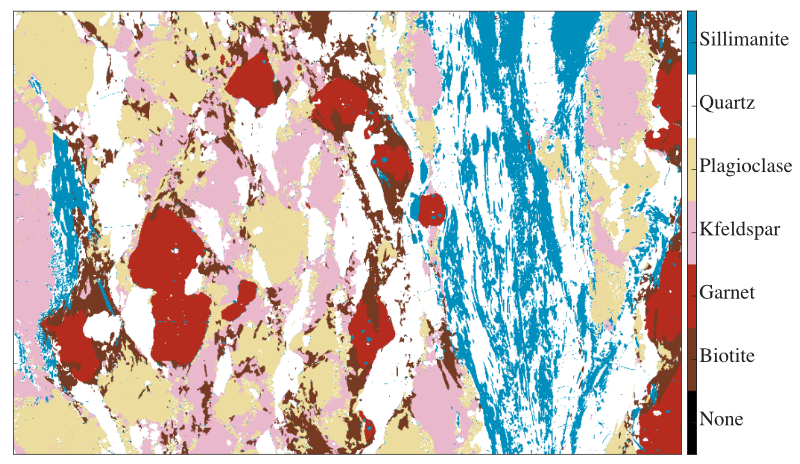
Figure: Example of mask image for a metapelite from the Himalaya published in Lanari & Duesterhoeft (2019). Each feature (mineral) is shown with a colour. Note that all the pixels of this image have been classified.
Classification parameters & algorithms
These tools are used to select an algorithm and maps to be used for classification.
Chemical system: how to add/remove maps?
The goal is to set the map input of the classification function. It is not required to use all available maps for classification; maps containing only noise are usually excluded.
The list of maps used by the classification function is displayed in the text field. The button Add Maps for Classification adds all maps available in the intensity tab of the primary menu. The button Edit Selected Map can have two modes: add (plus icon) or eliminate (minus icon) depending on whether the map selected in the primary menu is already available in the list or not. Clicking "plus" adds the selected map, whereas clicking "minus" eliminates the selected map from the list.
Algorithm selection
The machine learning algorithm used for classification can be selected via the algorithm menu available in the section Classification Parameters.
The following algorithms are available:
- Random Forest: An ensemble learning method for classification constructing a multitude of decision trees during training. The output of the random forest is the class selected by most trees (majority vote).
- Discriminant Analysis: Classification method that assumes that different classes generate data based on different Gaussian distributions. To train a classifier, it estimates the parameters of a Gaussian distribution for each class.
- Naive Bayes: Classification algorithm applying density estimation to the data and generating a probability model. The decision rule is based on the Bayes theorem.
- Support Vector Machine: Data points (p-dimensional vector) are separated into n classes by separating them with a (p-1)-dimensional hyperplane. The algorithm chooses the hyperplane so that the distance from it to the nearest data point on each side is maximised.
- Classification Tree: A decision tree is used as predictive model to classify the input features into classes via a series of decision nodes. Each leaf of the tree is labelled with a class.
- k-Nearest Neighbour: An object is classified by a plurality vote of its neighbours, with the object being assigned to the class most common among its k nearest neighbours.
- k-Means: Classification method that aims to partition n observations into k clusters in which each observation belongs to the cluster with the nearest cluster centroid, serving as a prototype of the cluster.
Principal component analysis (PCA) — Optional
The principal components of a collection of points in a real coordinate space are a sequence of vectors consisting of best-fitting lines, each of them defined as one that minimizes the average squared distance from the points to the line. These directions constitute an orthonormal basis in which different individual dimensions of the data are linearly uncorrelated. The first principal component can equivalently be defined as a direction that maximizes the variance of the projected data. Principal component analysis (PCA) is the process of computing the principal components and using them to perform a change of basis on the data.
The button Generate Maps of the Principal Components (PCA) generates a map for each principal component and stores them in the section Other of the primary menu.
If the tick-box incl. PCA is selected, the maps of principal components are included as additional dimensions for the classification. Example: if 8 intensity maps are considered, a total of 14 maps of PC are added to the classification input, 7 for a normal PCA and 7 for a normalised PCA.
Training and classification
A training set must be selected in the secondary menu in order to activate the classification button.
The button Classify (Train a Classifier & Classify) trains a new classifier and performs the classification using the algorithm selected in the menu and the specified set of maps.
A new figure containing up to four plots will open and be continuously updated during classification. Do not close this figure until the classification is complete, otherwise the plots will not be displayed.
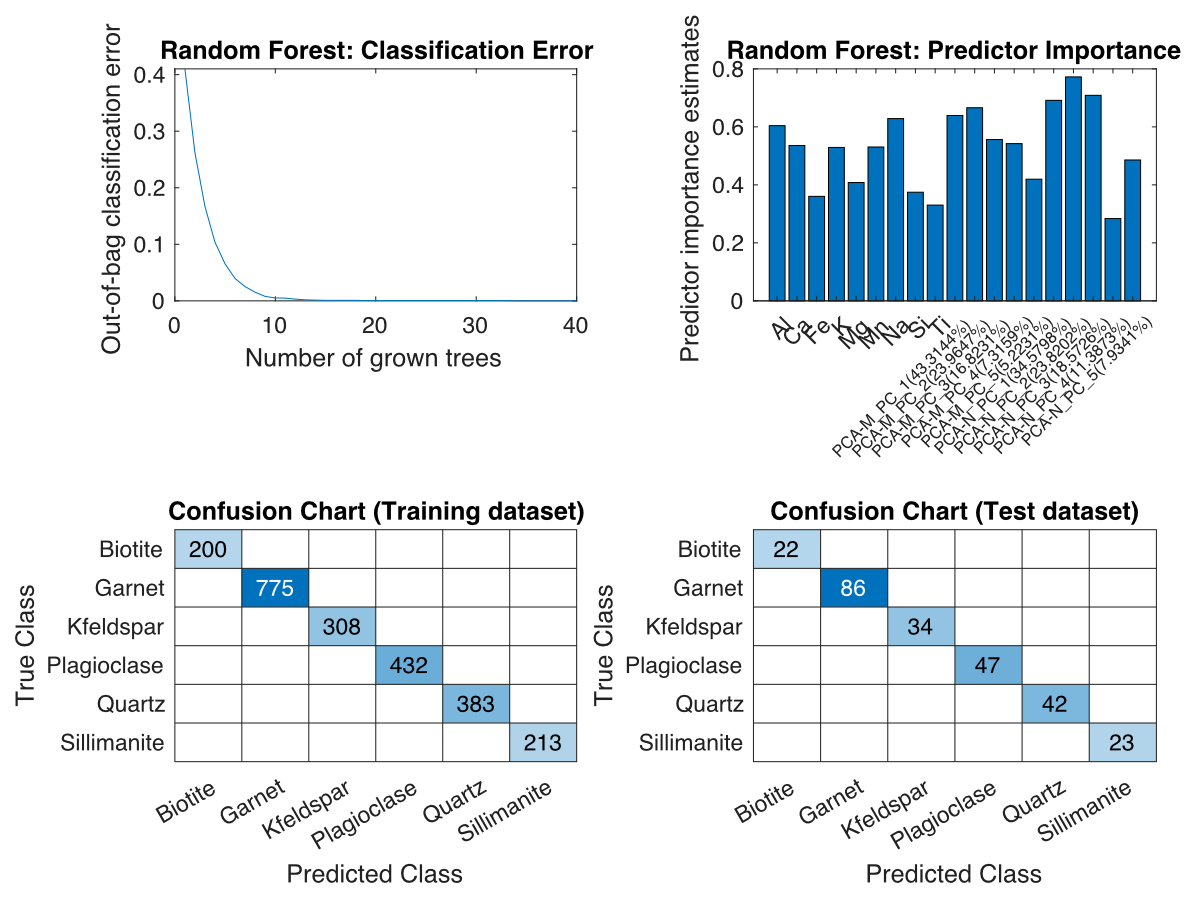
Figure: Plots for classification using the Random Forest algorithm. Top left: out-of-bag classification error vs. number of trees grown. Top right: Predictor importance. Bottom left: Confusion map of the training data set. Bottom right: Confusion plot of the test dataset.
Once the classification is achieved, a new mask file is generated and stored under Mask files in the secondary menu. The mask file is automatically selected and the mask image displayed in the main figure.
Filtering options
The following options apply to a given mask file. Select a mask file in the secondary menu to apply changes.
Select the option to hide pixels having a class probability lower than a given threshold for the selected mask file. Set the probability threshold used for hiding pixels. This value should range between 0 and 1.
The button Create new mask file with pixels filtered by probability generates a new mask file after filtering. This option replaces the BCR correction available in XMapTools 3 and is more adequate as only the misclassified pixels are excluded.
Mask analysis & visualisation
The modes of each class within a given region-of-interest can be exported using the tools provided in this section. Modes are given as surface percentage calculated using the number of pixels of each class. For minerals, this can be eventually extrapolated to volume fraction as discussed in Lanari & Engi (2017).
Select the ROI shape to be used to extract the local modes from the dropdown menu.
The button Add ROI allows a new region-of-interest (ROI) to be drawn on the figure. Note that a mask file must be selected before activating this mode. Results are displayed in the right window as a table containing the modes and number of pixels for each class and as a pie diagram.
The following ROI shapes are available:
- Rectangle: click to select the first corner and drag the mouse to the opposite corner defining a rectangle
- Polygon: click successively on the image to draw a polygon; right-clicking validates and closes automatically the shape
The button Plot Compositions generates a plot using the data selected in the primary menu (either intensity, or a merged map) and the mask file selected in the secondary menu.
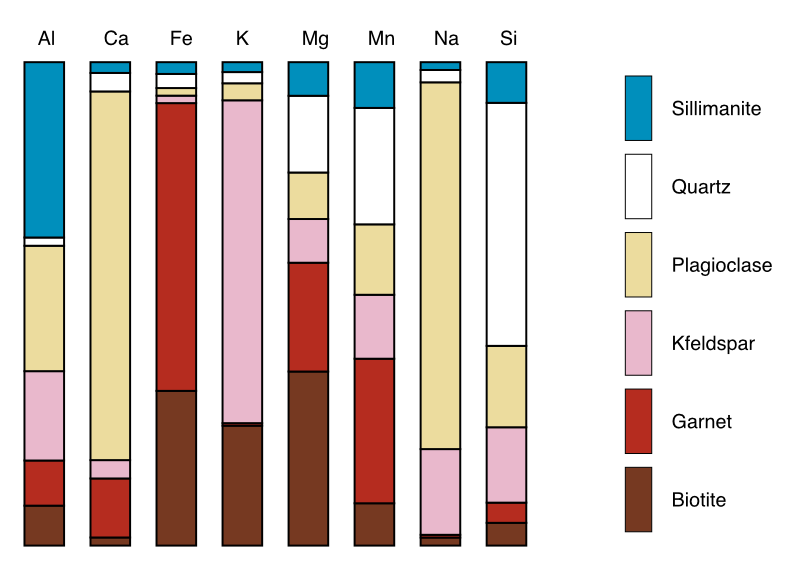
Figure: Example of compositional plot generated using a merged map (expressed in oxide wt%) and a mask file.
Calibration (EPMA)
The following steps are required to convert raw data (e.g. X-ray maps) into maps of chemical composition:
- Import standard data as spot analyses for the calibration of EPMA data
- Check/adjust/add standard data
- Calibrate using the Calibration Assistant for EPMA data
Standard data
The spot analyses used as internal standards in the calibration of EPMA data are referred to as "standards" in the following as they permit to define a calibration curve that correlates X-ray intensities to composition (e.g. expressed in oxide/element wt%).
It is necessary to import the standards from a standard file, check their positions and eventually correct, check the chemical compositions and eventually create new standards. All these steps can be achieved in XMapTools 4.
File Standards.txt
The file Standards.txt contains (i) the map coordinates and (ii) the spot analyses used for the standardisation. The map coordinates must be listed within a single row below the keyword >1. The oxide order is set below the keyword >2. X and Y must be the two last labels and must be listed in this specific order. The internal standards analyses are listed below the keyword >3 corresponding to the oxide order defined above (keyword >2).
>1 Here paste the image coordinates (Xmin Xmax Ymax Ymin)
56.739 57.239 43.691 43.371
>2 Here define the oxides order
SiO2 MgO FeO Al2O3 X Y
>3 Here paste the analyses
25.4800 11.260 29.050 21.1400 1.4800 68.310 39.999
52.9400 3.5300 3.0200 24.2300 0.0197 68.310 39.535
52.5800 3.6300 2.7900 24.7200 0.0195 68.331 39.511Import standards
The button Import (Import spots for Standards (from file)) is used to import standards from a file.
The box Import from Standards.txt is selected by default allowing the file 'Standards.txt' to be read automatically. If the file containing the standard data has a different name, unselect the box and it will be possible to select a file in the Pick a file pop-up window. At the moment all standards need to be stored within a single file. It is not possible to import standards from different files as existing standards will be eliminated when a new file is loaded.
Once loaded, standards are displayed on the main map with a label including the spot number and several plots are produced and shown in the category "Standards" of the live display module. You can get this global visualisation at any stage by selecting Standards (Spots) in the Secondary Menu. Three plots are produced if an element is selected in the primary menu, from top to bottom: (1) a plot showing intensity/composition versus sequence of standard to visualise if there is a good match between the standard compositions and the intensity values of the matching pixels; (2 and 3) two correlation maps, one for the selected element in the primary menu and a second one considering all elements.
Adjusting standard positions
To adjust the positions, display the map of a diagnostic element (Intensity) and select Standards (Spots) in the secondary menu. The two correlation maps should show a maximum value in yellow and the blue spot representing the current position should be centred on this optimum.
To adjust the position of the standards (all at once), put the mouse cursor over the blue circle showing up a transparent circle. Click on it and move the blue circle to the new position. The values in the two white fields on top will be adjusted. Then click on the button Refresh (important) to update the standard positions. The Refresh button is only available when standard positions have been changed and need to be saved.
Note: If no good correlation exists for a given element, the higher value of the second figure could not represent the optimal position.
Adding new standard point(s)
It is possible to add new standard points directly in XMapTools. Note that these will not be saved to the file Standards.txt, and if the file is loaded again all changes will be lost.
The button Add standard point adds a new standard at selected coordinates, set by clicking on the map after pressing the button. Compositional data can be filled directly in the table, when this standard is selected.
Procedure:
- Select an element map (e.g. SiO₂ for adding a spot of quartz) and select the item Standards (Spots) in the secondary menu
- Press the button Add Standard Point
- Click on the map at the position to add the new standard point
- (Optional) It is possible to adjust the position of any manual standard by moving the blue spot (transparent circle) or its label (cross-shaped cursor)
- The new standard point is automatically selected in the Secondary Menu and the composition table shown on the right side
- You can enter manually the oxide (wt%) composition of the new standard in the column 'wt%' in blue. The X-ray intensity of the corresponding pixel is already available in the 'Int' column
- There is nothing more to do — all data are automatically saved
- (Optional) You can rename any standard by double clicking on its name in the secondary menu
Map calibration
A calibration step, also known as standardisation, is required to convert intensity maps into compositional maps (Lanari et al. 2019).
All minerals/objects are calibrated at the same time in XMapTools 4. Therefore it is required to select a Mask File in the Secondary Menu to activate the button Calibrate.
The approach implemented in XMapTools 4 provides a module for auto multi-phase calibration. The button Calibrate opens the Calibration Assistant for EPMA Data. This button is only available when a mask file is selected in the Secondary Menu.
For more detailed information on the Calibration Assistant, refer to the embedded documentation accessible from the assistant.
Calibration assistant (EPMA)
The new approach implemented in XMapTools 4 provides a module for automatic multi-phase calibration. The general procedure is described in De Andrade et al. (2006) and an advanced approach including pseudo-background correction is described in Lanari et al. (2019).
Strategy: advantages and pitfalls
An automatic calibration is performed taking into account all spot analyses and all masks when the Calibrate button is pressed. The new algorithm first performs a general fit including all standards and then adjusts the calibration for each mineral. All calibration curves, including those for the general fit, are accessible from the tree menu.
When you press the Apply Standardisation button, all the calibrated maps for each mineral as well as a merged map are created and sent back to XMapTools.
Important
First make sure you check the quality of the calibration curves generated by the auto function!
The automatic function will work if all minerals have been measured with at least a few spot analyses and if there is at least one mineral with a composition above 1 wt% for each element. If a mineral or other feature (e.g. fracture) has no spot analyses, the program extrapolates a calibration from the general fit and thus "predicts" a composition. At this stage this composition is likely to be off because matrix effects are ignored!
INFO
- If you close the Calibration Assistant window, no calibrated data or calibration settings will be saved.
- When you press Apply Standardisation, all maps are sent to XMapTools.
Displaying calibration curves
The Calibration Assistant for EPMA Data opens when the Calibrate button in XMapTools is pressed.
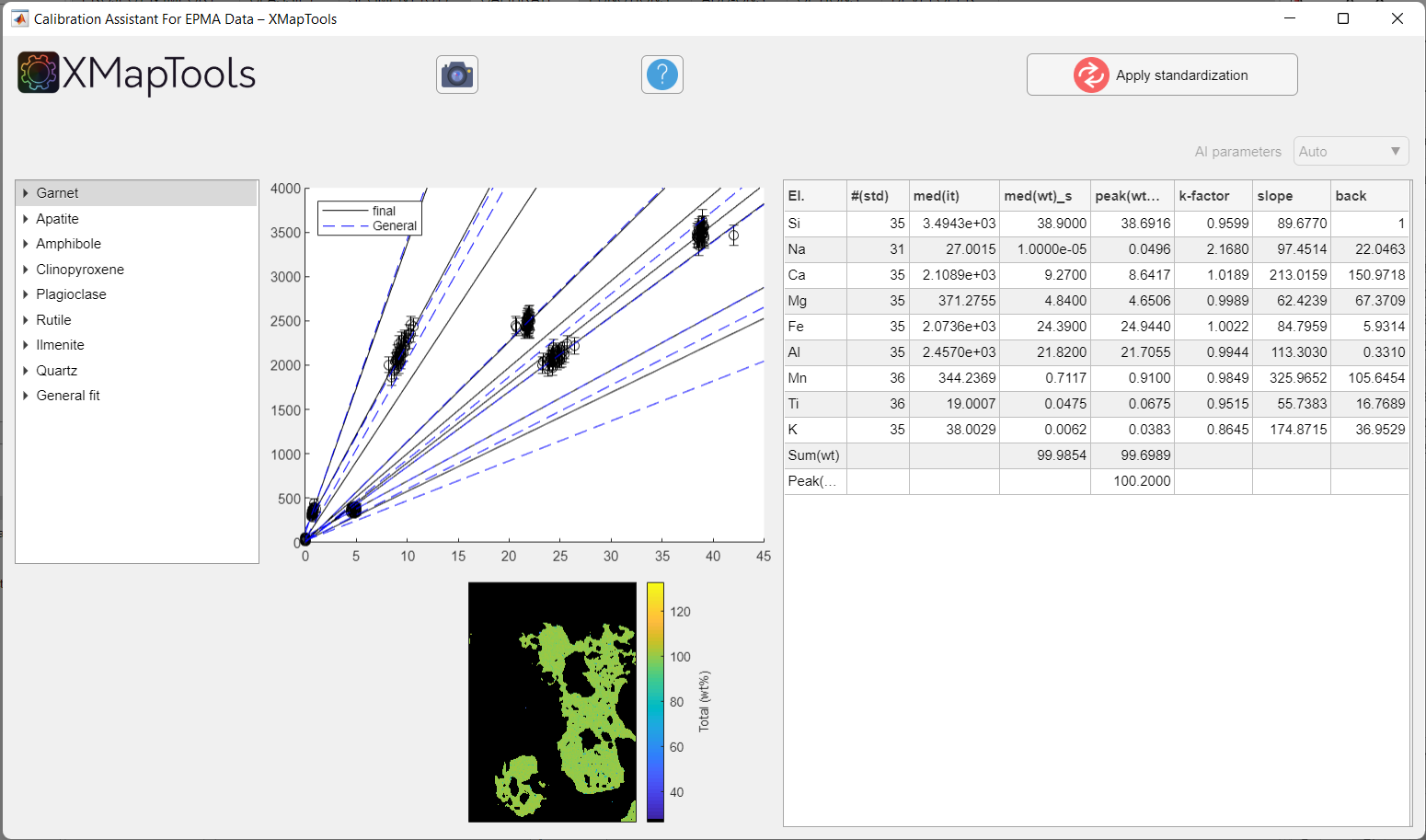
Figure: Example calibration for a clinopyroxene-garnet amphibolite metapelite of the Brasília orogen (Brazil), published in Tedeschi et al. (2017). Note that the calibration curves for all minerals are displayed when the window pops up.
Use the tree menu on the left to navigate through the list of minerals and elements and to view calibration curves.
When you select a mineral from the tree menu, a single plot showing the sum of elements/oxides (total wt%) is displayed. The plot in the middle shows all calibration curves, for all elements of the selected mineral. Some data is displayed in a table:
| Column | Description |
|---|---|
| El. | Element name (of the map); includes sum(wt%) and Peak(SumOx) labels |
| #(std) | Number of internal standards (spot analyses) used to calibrate the phase |
| med(it) | Median intensity value for all pixels of the selected mineral for each element |
| med(wt)_s | Median composition of all internal standard measurements (spot analyses) |
| mode(wt)_m | Most common composition in the calibrated pixels |
| k factor | Difference between the general fit calibration and the final mineral calibration (1 = identical) |
| Slope | Slope of the calibration curve |
| Background | Intercept of the calibration curve |
TIP
The values of mode(SumOx) and Sum(wt) may be different, in which case the median is likely to be influenced by non-Gaussian signals and may not be comparable with the median of the spot analyses. The comparison of both columns can be used to detect potential calibration problems.
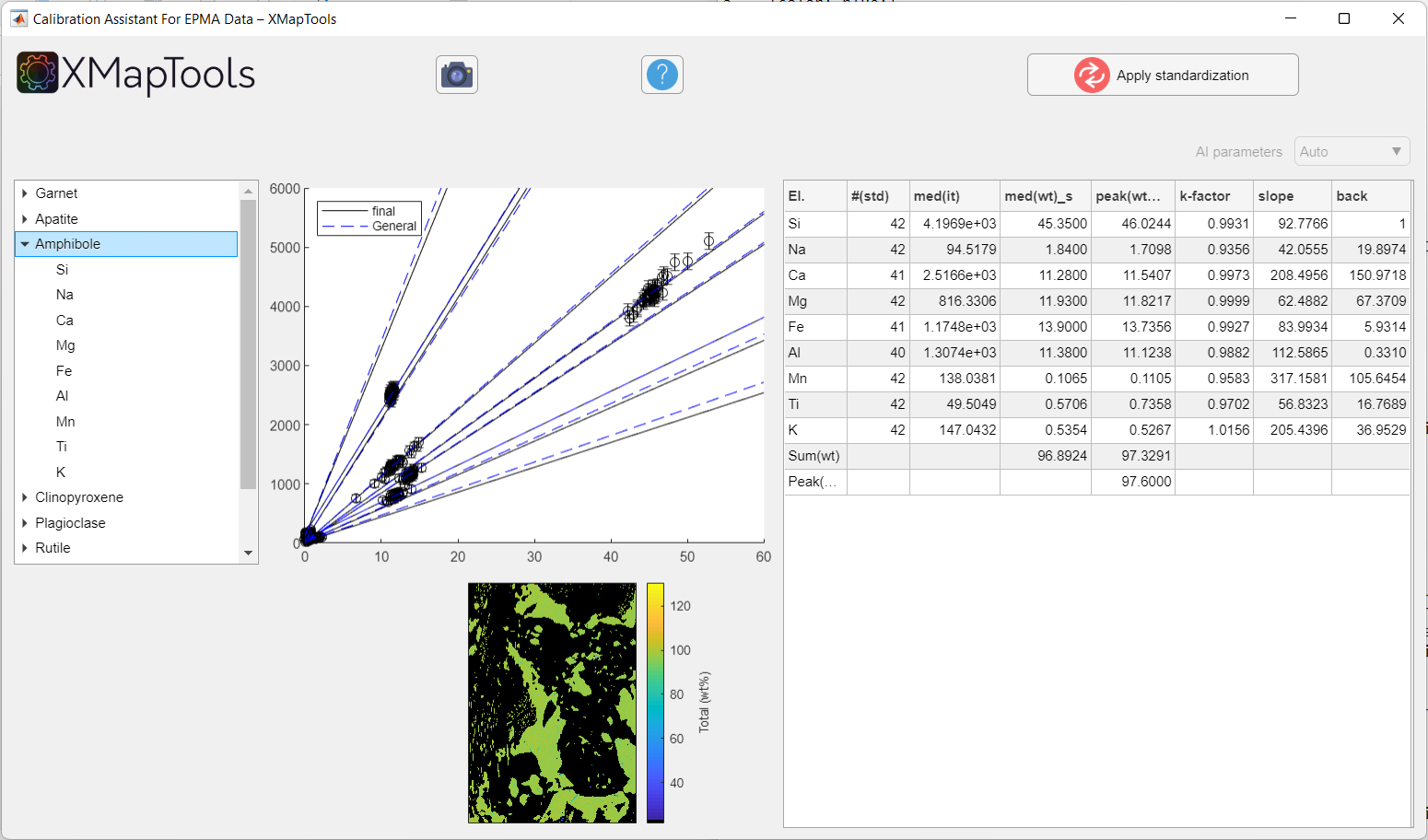
Figure: Example of calibration when a mineral is selected. All calibration curves for a given mineral are displayed.
To view a specific calibration curve (for a particular element), expand the menu by clicking on the small arrow to the left of the mineral name and select an element. The corresponding calibration curve is displayed together with the corresponding quantitative map (oxide wt-%).
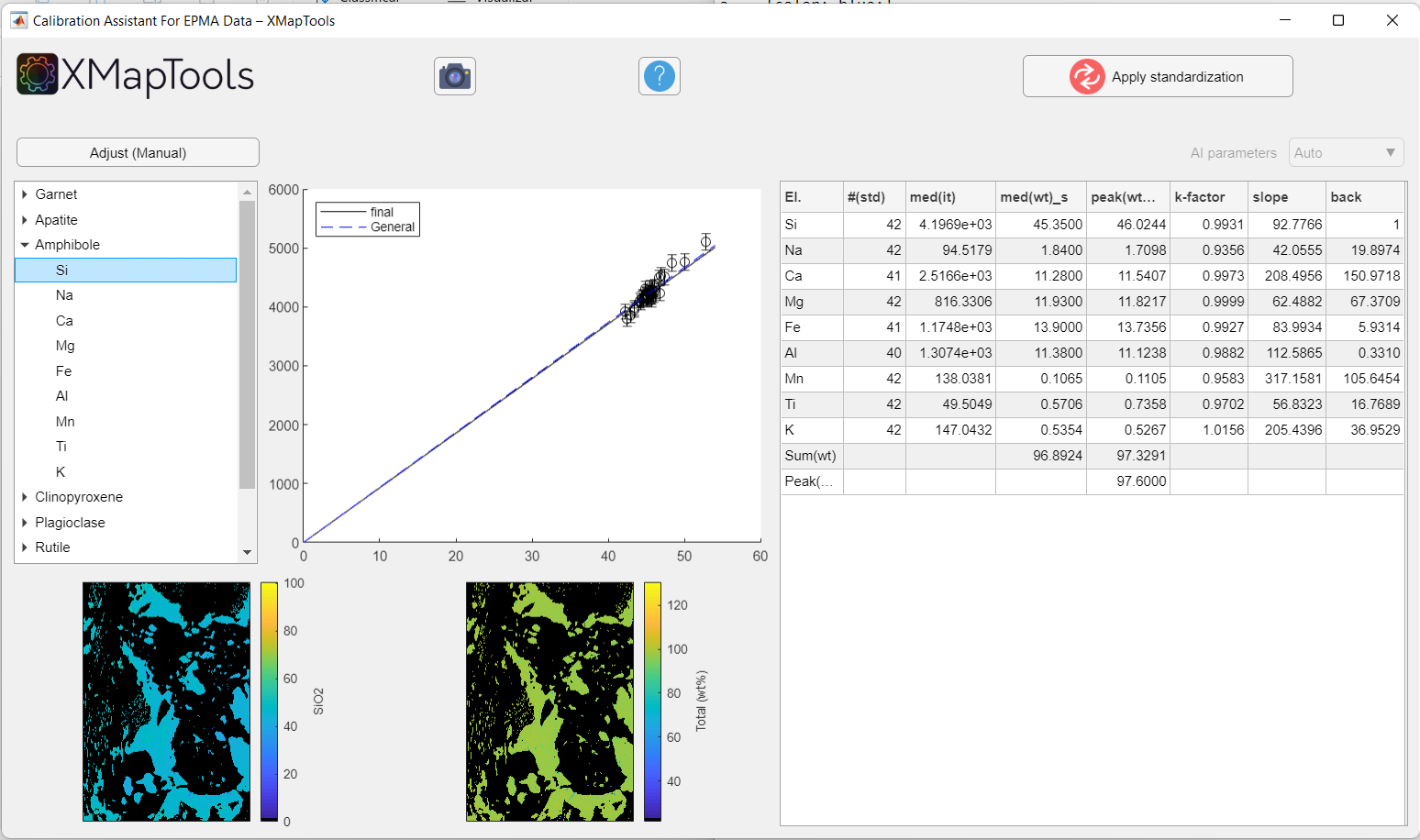
Figure: Example when an element of a given mineral phase is selected. The calibration curve and quantitative map for a given element are displayed.
Adjusting a calibration curve
When an element is selected, the Adjust ![]() button appears above the tree menu.
button appears above the tree menu.
Clicking this button displays two fields containing the values for background and slope: ![]()
Values can be changed manually by entering new values in the appropriate field. Press Enter to calculate and display the new calibration curve on the graph (this operation may take a few seconds).
Displaying results of the general fit
Selecting General Fit (last option in the tree menu) will display a plot of the calibration curves for all elements. It is not possible to adjust the calibration curves in the general fit. This fit is automatically performed first by the program and is no longer used once the calibration of each mineral has been achieved.
Apply calibration
After checking each calibration curve and adjusting if necessary, use the Apply Standardisation button ![]() to generate the calibrated maps.
to generate the calibrated maps.
By applying the standardisation:
- The Quanti option in the Primary Menu becomes available, where quantitative maps in element/oxide wt-% of each phase can be displayed.
- The Merged option in the Primary Menu also becomes available. A set of merged maps (i.e. quantitative maps in oxide wt-% for all phases considered together) is automatically generated.
Notes
- Red dots indicate outliers that were not included in the calculation of the calibration curves.
- Moving the cursor over the images brings up a Image Menu at the top right. This menu includes options to zoom, save and copy the images.
Local bulk compositions
A local bulk composition (abbreviation: LBC) represents the bulk composition of a spatial domain in a rock determined by integrating pixel compositions. As discussed in Lanari and Engi (2017), it is necessary to apply a density correction prior to exporting any local bulk composition.
To export local bulk compositions the following steps should be employed:
- Generate a density map
- Generate a merged map (if not available yet)
- Select an area of interest to extract the local bulk composition
Generate a density map
A density map is a map containing density data for each pixel of a map. It is calculated for a given mask file. Select a mask file in the secondary menu.
The button Generate Density Map (from a mask file) allows a density map to be generated from the selected mask file. A mask file should be selected to activate the button. Pressing this button opens a window with predefined average density values (provided that the mineral name was recognised and a reference value available in the internal database; when full names of minerals in English are used, the mineral should be recognised).
Note: mineral density values can be obtained from the website webmineral.com.
A density map will be created and stored under the category Other in the Primary Menu with the name 'Density [maskfile_name]'.
Generate a merged map
Merged maps are maps for which all pixels hold a chemical composition. A merged map is automatically created by the standardisation function in XMapTools 4. If you need to create a merged map manually, follow the procedure below.
In the primary menu, unfold Quanti and select a quanti map (mineral). The button Merge (Merge Quanti Data) in the section Calibrate becomes available. In the window that pops up, select the quanti maps to be merged. Select 'Ok' to generate the merged maps that will be stored in the category Merged.
Select ROI and calculate LBC
A local bulk composition can be calculated from a region-of-interest (ROI). The ROI can be a rectangle or a polygon. The density correction is automatically applied.
Select a merged map in the primary menu and display an element. In the dropdown menu, select the wanted ROI shape: Rectangle ROI or Polygon ROI.
The button Add a ROI for LBC extraction allows a region-of-interest (ROI) to be drawn on the figure. If Rectangle ROI is selected, click on a corner, hold and drag to draw a rectangle. If Polygon ROI is selected, click on the image to draw the polygon and close by selecting the first point or right-clicking.
When a ROI is available, the table containing the local bulk composition appears in the live display module. The first column shows the element list, whereas compositions are listed in the column composition (unit: wt%). Below the table, a pie chart shows the repartition of the elements/oxides by weight.
The ROI can be edited and the composition values in the table and the pie chart are automatically updated.
The density map used is shown in a dropdown menu located in the live display module.
The button Copy Data to clipboard may be used to copy the LBC data from the table. The button Save may be used to save the LBC data as a .txt file.
Approximation of uncertainties for LBC
An uncertainty approximation similar to what is described in Lanari and Engi (2017) is available.
Select a merged map and create a Rectangle ROI. Define the number of simulations Sim (default 100) and the shift Px in pixel (default 20). The shape will be randomly displaced and resized using two random variables calculated from the shift value (assuming a Gaussian distribution and the value of Px as 1 sigma expressed in number of pixels). Click on the button Calculate uncertainties using Monte-Carlo.
The areas used to approximate an uncertainty are plotted in a new figure and the result is shown in the table of the live display module. The column composition shows the original local bulk composition (selected ROI). The column 2std shows the 2 standard deviation value (note that the distributions are usually Gaussian as shown by Lanari & Engi (2017)). The last column shows the mean value of all compositions. This value should match the composition of the original ROI. If not, this means that a Gaussian distribution cannot be assumed for this element and the uncertainty is not correct.
References
- De Andrade, V., Vidal, O., Lewin, E., O'Brien, P., Agard, P. (2006). Quantification of electron microprobe compositional maps of rock thin sections: an optimized method and examples. Journal of Metamorphic Geology, 24, 655–668.
- Lanari, P., & Engi, M. (2017). Local bulk composition effects on metamorphic mineral assemblages, Reviews in Mineralogy and Geochemistry, 83, 55–102.
- Lanari, P., Vho, A., Bovay, T., Airaghi, L., Centrella, S. (2019). Quantitative compositional mapping of mineral phases by electron probe micro-analyser. Geological Society of London, Special Publications, 478, 39–63.
